Found 2 results
Open Access
Article
04 June 2025Modular Transcriptional Regulation Using Switchable Transcription Terminators and Aptamers: Design and Optimization in Synthetic Biology
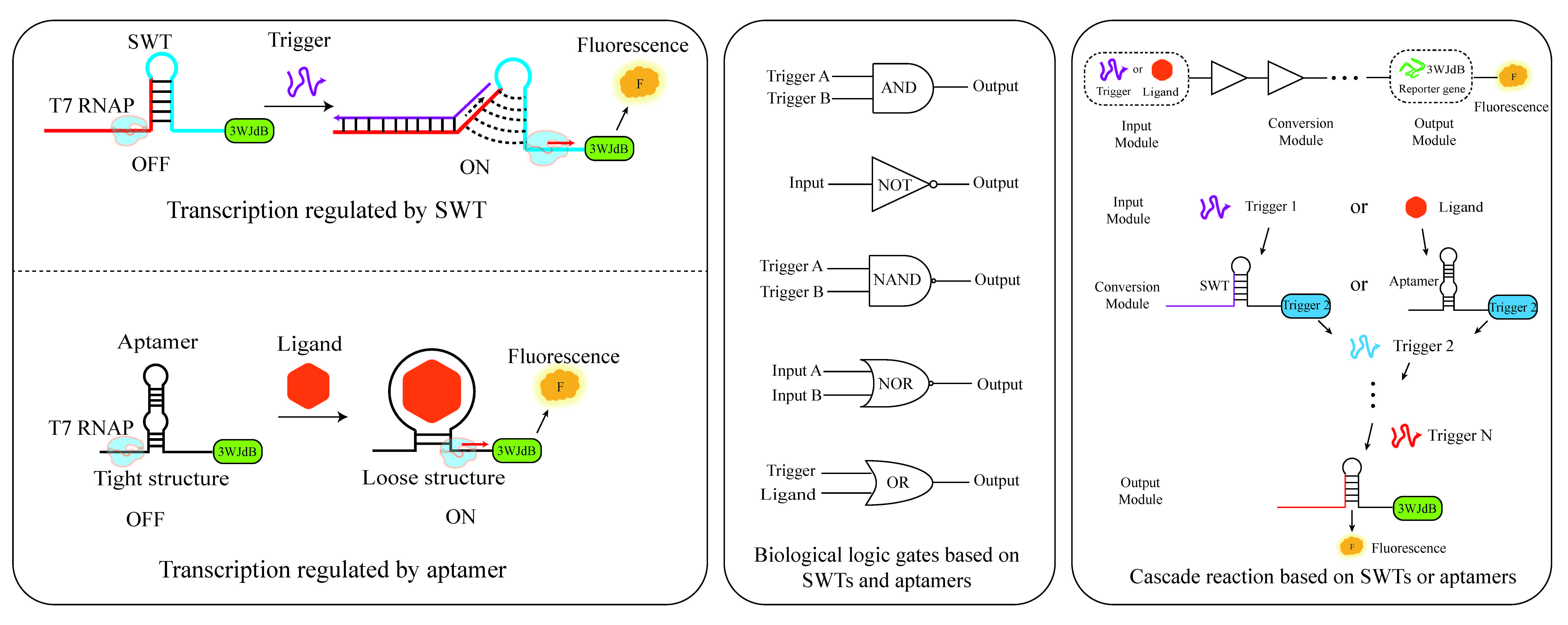
Transcriptional regulation is a key step in gene expression control. While transcription factor-based regulation has been widely used and offers robust control over gene expression, it can sometimes face challenges such as achieving high specificity, rapid dynamic responses, and fine-tuned regulatory precision, which have motivated the exploration of alternative regulatory strategies. With the development of synthetic biology, novel genetic elements such as Switchable Transcription Terminators (SWT) and aptamers provide more flexible and programmable strategies for transcriptional regulation. However, the independent regulatory capabilities of these two types of elements and their combined regulatory mechanisms still require further investigation. In this study, based on an in vitro transcription system, we systematically explored the transcriptional regulation potential of SWT and aptamers. We innovatively combined these two elements to construct a modular gene expression regulation system. First, we screened and optimized a series of SWTs, obtaining high-performance SWTs with low leakage expression and high ON/OFF ratios. These were further validated for reproducibility of their regulatory performance in E. coli. Next, we constructed multi-level cascading circuits using SWTs, successfully extending the system to six levels and building four types of biological logic gates based on SWT in vitro: AND gate, NOT gate, NAND gate, and NOR gate. Furthermore, based on a previously identified thrombin aptamer capable of transcriptional regulation, we confirmed that ligand binding significantly promoted gene transcription. Finally, we integrate switchable transcription terminators (SWTs) and aptamers to create a modular, ligand-responsive system. We combined aptamers with SWTs to construct heterologous input logic gates, successfully improving the precision and dynamic range of regulation. Compared to the individual regulation of SWT and aptamer, the Aptamer-SWT synergistic regulation enhanced transcription activation by up to 3.3-fold and 7.84-fold, respectively. Additionally, we co-utilized these two genetic elements to construct heterologous input AND and OR gates in vitro. This study expands the strategies for gene expression regulation and provides new elements and theoretical support for efficient, programmable transcriptional regulation in synthetic biology. This system holds potential for biosensing, gene circuit design, and nucleic acid therapy applications.
Open Access
Review
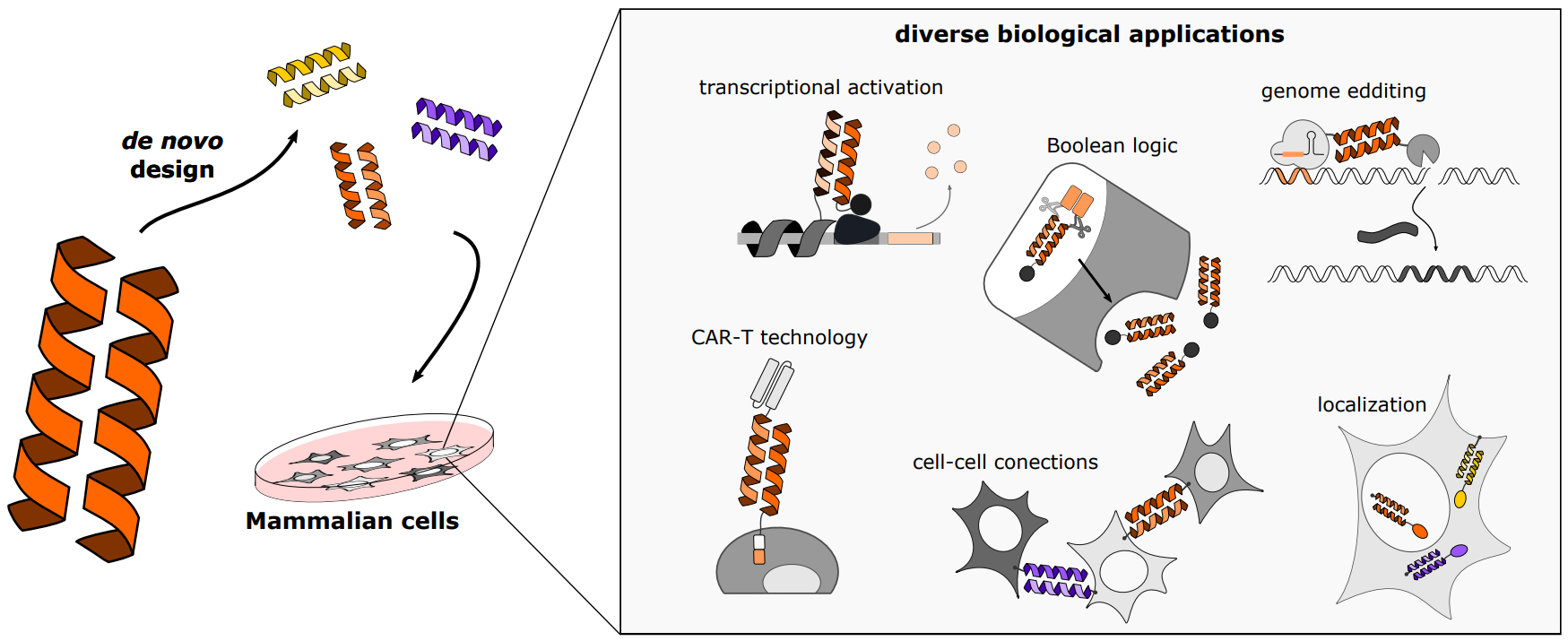
06 April 2023Coiled Coils as Versatile Modules for Mammalian Cell Regulation
Synthetic biology is a rapidly growing field that allows us to better understand biological processes at the molecular level, and enables therapeutic interventions and biotechnological applications. One of the most powerful tools in synthetic biology is the small, customizable, and modular protein–protein interaction domains, which is used to regulate a wide variety of processes within mammalian cells. Here we review designed coiled coil dimers that represent a set of heterodimerization domains with many advantages. These dimers have been useful for directing the localization of selected proteins within cells, enhancing chemical or light-regulated transcription, creating fast proteolysis-based responsive systems and protein secretion, genome editing, and cell–cell interaction motifs. Additionally, we will discuss how these building blocks are used in diverse applications, such as CAR T cell regulation and genome editing. Finally, we will look at the potential for future advances in synthetic biology using these building modules.